Combining measurements#
When we do a fit, we can have additional knowledge about a parameter from other measurements. This can be taken into account either through a simultaneous fit or by adding a constraint (subsidiary measurement).
Adding a constraint#
If we know a parameters value from a different measurement and want to constraint this using its uncertainty, a Gaussian constraint can be added to the likelihood as
In general, additional terms can be added to the likelihood arbitrarily in zfit, be it to incorporate other shaped measurements or to add penalty terms to confine a fit within boundaries.
import hepunits as u
import matplotlib.pyplot as plt
import mplhep
import numpy as np
import particle.literals as lp
import tensorflow as tf
import zfit.z.numpy
plt.rcParams['figure.figsize'] = (8, 6)
mu_true = lp.B_plus.mass * u.MeV
sigma_true = 50 * u.MeV
# number of signal and background
n_sig_rare = 120
n_bkg_rare = 700
# create some data
signal_np = np.random.normal(loc=mu_true, scale=sigma_true, size=n_sig_rare)
bkg_np_raw = np.random.exponential(size=20000, scale=700)
bkg_np = bkg_np_raw[bkg_np_raw < 1000][:n_bkg_rare] + 5000 # just cutting right, but zfit could also cut
# Firstly, the observable and its range is defined
obs = zfit.Space('Bmass', 5000, 6000) # for whole range
# load data into zfit and let zfit concatenate the data
signal_data = zfit.Data(signal_np, obs=obs)
bkg_data = zfit.Data(bkg_np, obs=obs)
data = zfit.data.concat([signal_data, bkg_data])
# (we could also do it manually)
# data = zfit.Data(array=np.concatenate([signal_np, bkg_np], axis=0), obs=obs)
# Parameters are specified: (name (unique), initial, lower, upper) whereas lower, upper are optional
mu = zfit.Parameter('mu', 5279, 5100, 5400)
sigma = zfit.Parameter('sigma', 20, 1, 200)
sig_yield = zfit.Parameter('sig_yield', n_sig_rare + 30,
step_size=3) # step size: default is small, use appropriate
signal = zfit.pdf.Gauss(mu=mu, sigma=sigma, obs=obs, extended=sig_yield)
lam = zfit.Parameter('lambda', -0.002, -0.1, -0.00001, step_size=0.001) # floating, also without limits
bkg_yield = zfit.Parameter('bkg_yield', n_bkg_rare - 40, step_size=1)
comb_bkg = zfit.pdf.Exponential(lam, obs=obs, extended=bkg_yield)
# The final model is the combination of the signal and backgrond PDF
model = zfit.pdf.SumPDF([comb_bkg, signal])
constraint = zfit.constraint.GaussianConstraint(mu, observation=5275 * u.MeV, sigma=15 * u.MeV)
nll = zfit.loss.ExtendedUnbinnedNLL(model, data, constraints=constraint)
minimizer = zfit.minimize.Minuit(gradient="zfit")
result = minimizer.minimize(nll)
result.hesse();
print(result)
FitResult
of
<ExtendedUnbinnedNLL model=[<zfit.<class 'zfit.models.functor.SumPDF'> params=[Composed_autoparam_1, Composed_autoparam_2]] data=[<zfit.Data: Data obs=('Bmass',) shape=(820, 1)>] constraints=[<zfit.core.constraint.GaussianConstraint object at 0x7fca16e6b190>]>
with
<Minuit Minuit tol=0.001>
╒═════════╤═════════════╤══════════════════╤═════════╤══════════════════════════════╕
│ valid │ converged │ param at limit │ edm │ approx. fmin (full | opt.) │
╞═════════╪═════════════╪══════════════════╪═════════╪══════════════════════════════╡
│
True
│ True
│ False
│ 0.00018 │ 866.35 | 9979.792 │
╘═════════╧═════════════╧══════════════════╧═════════╧══════════════════════════════╛
Parameters
name value (rounded) hesse at limit
--------- ------------------ ----------- ----------
bkg_yield 729.034 +/- 32 False
sig_yield 91.117 +/- 20 False
lambda -0.0016528 +/- 0.00014 False
mu 5284.01 +/- 7.6 False
sigma 39.4786 +/- 9.2 False
Simultaneous fits#
Sometimes, we don’t want to fit a single channel but multiple with the same likelihood and having shared parameters between them. In this example, we will fit the decay simultaneously to its resonant control channel.
A simultaneous likelihood is the product of different likelihoods defined by
and becomes in the NLL a sum
n_sig_reso = 40000
n_bkg_reso = 3000
# create some data
signal_np_reso = np.random.normal(loc=mu_true, scale=sigma_true * 0.7, size=n_sig_reso)
bkg_np_raw_reso = np.random.exponential(size=20000, scale=900)
bkg_np_reso = bkg_np_raw_reso[bkg_np_raw_reso < 1000][:n_bkg_reso] + 5000
# load data into zfit
obs_reso = zfit.Space('Bmass_reso', 5000, 6000)
signal_data_reso = zfit.Data(signal_np_reso, obs=obs_reso)
bkg_data_reso = zfit.Data(bkg_np_reso, obs=obs_reso)
data_reso = zfit.data.concat([signal_data_reso, bkg_data_reso])
Plotting a simultaneous loss#
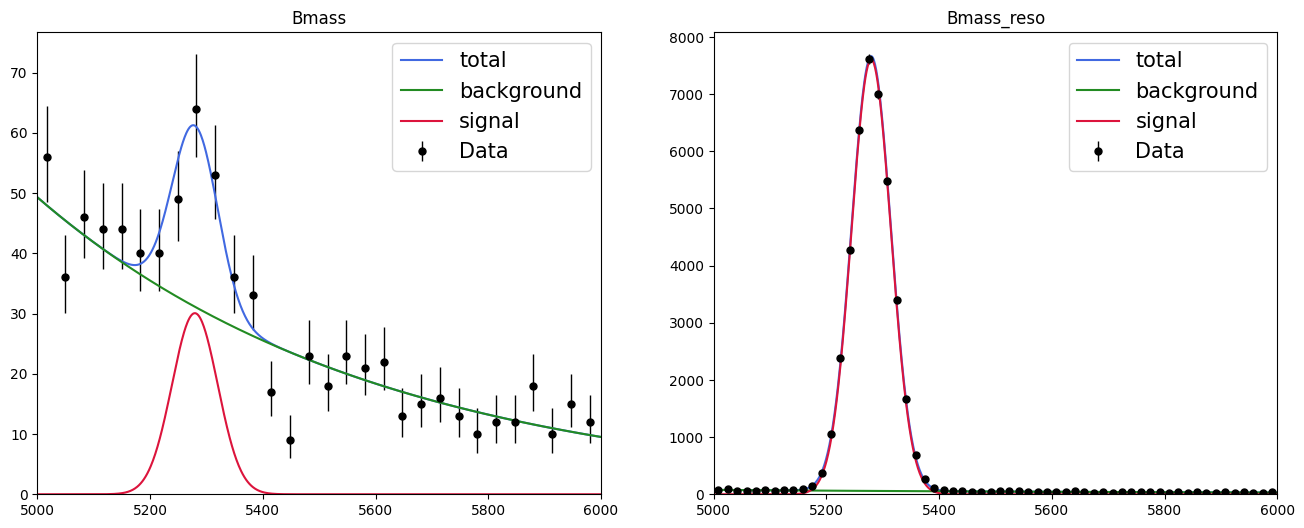
Since the definition of a simultaneous fit is as above, it is simple to plot each component separately: either my using the attributes of the loss to access the models and plot in a general fashion or directly reuse the model and data from before; we created them manually before.
# Sets the values of the parameters to the result of the simultaneous fit
# in case they were modified.
zfit.param.set_values(nll_simultaneous.get_params(), result_simultaneous)
def plot_fit_projection(model, data, nbins=30, ax=None):
# The function will be reused.
if ax is None:
ax = plt.gca()
lower, upper = data.data_range.limit1d
# Creates and histogram of the data and plots it with mplhep.
counts, bin_edges = np.histogram(data.unstack_x(), bins=nbins)
mplhep.histplot(counts, bins=bin_edges, histtype="errorbar", yerr=True,
label="Data", ax=ax, color="black")
binwidth = np.diff(bin_edges)[0]
x = tf.linspace(lower, upper, num=1000) # or np.linspace
# Line plots of the total pdf and the sub-pdfs.
y = model.ext_pdf(x) * binwidth
ax.plot(x, y, label="total", color="royalblue")
for m, l, c in zip(model.get_models(), ["background", "signal"], ["forestgreen", "crimson"]):
ym = m.ext_pdf(x) * binwidth
ax.plot(x, ym, label=l, color=c)
ax.set_title(data.data_range.obs[0])
ax.set_xlim(lower, upper)
ax.legend(fontsize=15)
return ax
fig, axs = plt.subplots(1, 2, figsize=(16, 6))
for mod, dat, ax, nb in zip(nll_simultaneous.model, nll_simultaneous.data, axs, [30, 60]):
plot_fit_projection(mod, dat, nbins=nb, ax=ax)
Discovery test#
We observed an excess of our signal:
print(result_simultaneous.params[sig_yield])
{'value': 91.95480704554072, 'hesse': {'error': 17.39989772395696, 'cl': 0.68268949}}
Now we would like to compute the significance of this observation or, in other words, the probabilty that this observation is the result of the statistical fluctuation. To do so we have to perform an hypothesis test where the null and alternative hypotheses are defined as:
\(H_{0}\), the null or background only hypothesis, i.e. \(N_{sig} = 0\);
\(H_{1}\), the alternative hypothesis, i.e \(N_{sig} = \hat{N}_{sig}\), where \(\hat{N}_{sig}\) is the fitted value of \(N_{sig}\) printed above.
In hepstats to formulate a hypothesis you have to use the POI (Parameter Of Interest) class.
from hepstats.hypotests.parameters import POI
# the null hypothesis
sig_yield_poi = POI(sig_yield, 0)
What the POI class does is to take as input a zfit.Parameter instance and a value corresponding to a given hypothesis. You can notice that we didn’t define here the alternative hypothesis as in the discovery test the value of POI for alternate is set to the best fit value.
The test statistic used is the profile likelihood ratio and defined as:
where \(\hat{\theta}\) are the best fitted values of the nuisances parameters (i.e. background yield, exponential slope…), while \(\hat{\hat{\theta}}\) are the fitted values of the nuisances when \({N}_{sig} = 0\).
From the test statistic distribution a p-value can computed as
where \(q_{0}^{obs}\) is the value of the test statistic evaluated on observed data.
The construction of the test statistic and the computation of the p-value is done in a Calculator object in hepstats. In this example we will use in this example the AsymptoticCalculator calculator which assumes that \(q_{0}\) follows a \(\chi^2(ndof=1)\) which simplifies the p-value computation to
The calculator objects takes as input the likelihood function and a minimizer to profile the likelihood.
from hepstats.hypotests.calculators import AsymptoticCalculator
# construction of the calculator instance
calculator = AsymptoticCalculator(input=nll_simultaneous, minimizer=minimizer)
calculator.bestfit = result_simultaneous
# equivalent to above
calculator = AsymptoticCalculator(input=result_simultaneous, minimizer=minimizer)
There is another calculator in hepstats called FrequentistCalculator which constructs the test statistic distribution \(f(q_{0} |H_{0})\) with pseudo-experiments (toys), but it takes more time.
The Discovery class is a high-level class that takes as input a calculator and a POI instance representing the null hypothesis, it basically asks the calculator to compute the p-value and also computes the signifance as
from hepstats.hypotests import Discovery
discovery = Discovery(calculator=calculator, poinull=sig_yield_poi)
discovery.result()
p_value for the Null hypothesis = 2.2687673961740984e-10
Significance (in units of sigma) = 6.2343169775013285
(2.2687673961740984e-10, 6.2343169775013285)
So we get a significance of about \(7\sigma\) which is well above the \(5 \sigma\) threshold for discoveries 😃.
Upper limit calculation#
Let’s try to compute the discovery significance with a lower number of generated signal events.
# Sets the values of the parameters to the result of the simultaneous fit
zfit.param.set_values(nll_simultaneous.get_params(), result_simultaneous)
sigma_scaling.floating = False
# Creates a sampler that will draw events from the model
sampler = model.create_sampler()
# Creates new simultaneous loss
nll_simultaneous_low_sig = zfit.loss.ExtendedUnbinnedNLL(model, sampler) + nll_reso
# Samples with sig_yield = 10. Since the model is extended the number of
# signal generated is drawn from a poisson distribution with lambda = 10.
sampler.resample({sig_yield: 10})
calculator_low_sig = AsymptoticCalculator(input=nll_simultaneous_low_sig, minimizer=minimizer)
discovery_low_sig = Discovery(calculator=calculator_low_sig, poinull=sig_yield_poi)
discovery_low_sig.result()
print(f"\n {calculator_low_sig.bestfit.params} \n")
Get fit best values!
W MnHesse Analytical calculator [ -1.69814812e-05 -2.025312624e-05 -0.0509158992 -0.1702872609 0.03751929868 -0.2524187955 0.01524917472 0.01348904981] numerical [ -1.726421058e-05 -3.2894097e-05 -0.05831339971 -0.1745895082 0.03253207527 -0.2524915913 0.01524631393 0.01039098984] g2 [ 0.001333849566 0.00689583839 8902.009533 661422.6047 253630.8392 146990.7316 78459.2199 2072.924895]
p_value for the Null hypothesis = 0.30336501365699386
Significance (in units of sigma) = 0.5147467113695439
name value (rounded) at limit
-------------- ------------------ ----------
bkg_yield 743.152 False
sig_yield 6.83483 False
lambda -0.00168844 False
mu 5279.42 False
sigma 40.5315 False
reso_bkg_yield 2960.89 False
reso_sig_yield 40039.1 False
lambda_reso -0.000955933 False
We might consider computing an upper limit on the signal yield instead. The test statistic for an upper limit calculation is
and the p-value is
The upper limit on \(N_{sig}\), \(N_{sig, \uparrow}\) is found for \(p_{N_{sig, \uparrow}} = 1 - \alpha\), \(\alpha\) being the confidence level (typically \(95 \%\)). The upper limit is found by interpolation of the p-values as a function of \(N_{sig}\), which is done the UpperLimit class. We have to give the range of values of \(N_{sig}\) to scan using the POIarray class which as the POI class takes as input the parameter but takes several values to evaluate the parameter instead of one.
from hepstats.hypotests import UpperLimit
from hepstats.hypotests.parameters import POIarray
#Background only hypothesis.
bkg_only = POI(sig_yield, 0)
# Range of Nsig values to scan.
sig_yield_scan = POIarray(sig_yield, np.linspace(0, 70, 10))
ul = UpperLimit(calculator=calculator_low_sig, poinull=sig_yield_scan, poialt=bkg_only)
ul.upperlimit(alpha=0.05);
Get fitted values of the nuisance parameters for the alternative hypothesis!
FitResult
of
<ExtendedBinnedNLL model=[<zfit.models.tobinned.BinnedFromUnbinnedPDF object at 0x7fc9fba32290>, <zfit.models.tobinned.BinnedFromUnbinnedPDF object at 0x7fc9fbb5aa10>] data=[<zfit._data.binneddatav1.BinnedData object at 0x7fca02302210>, <zfit._data.binneddatav1.BinnedData object at 0x7fc9fbf31b50>] constraints=[]>
with
<Minuit Minuit tol=0.001>
╒═════════╤═════════════╤══════════════════╤═════════╤══════════════════════════════╕
│ valid │ converged │ param at limit │ edm │ approx. fmin (full | opt.) │
╞═════════╪═════════════╪══════════════════╪═════════╪══════════════════════════════╡
│
True
│ True
│ False
│ 7.2e-05 │ -288679.11 | -287313.7 │
╘═════════╧═════════════╧══════════════════╧═════════╧══════════════════════════════╛
Parameters
name value (rounded) at limit
-------------- ------------------ ----------
bkg_yield 749.943 False
lambda -0.00169807 False
mu 5279.42 False
sigma 40.5415 False
reso_bkg_yield 2962.38 False
reso_sig_yield 40038 False
lambda_reso -0.0009551 False
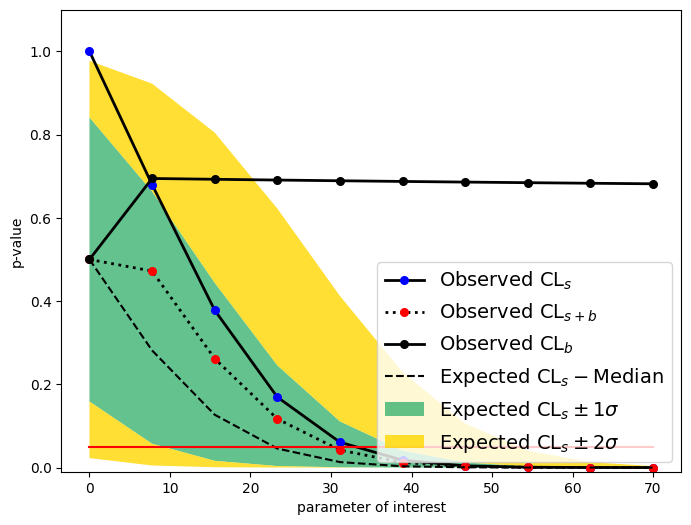
Observed upper limit: sig_yield = 32.43994223209022
Expected upper limit: sig_yield = 27.26225533489645
Expected upper limit +1 sigma: sig_yield = 38.66791298913074
Expected upper limit -1 sigma: sig_yield = 19.34122302338897
Expected upper limit +2 sigma: sig_yield = 53.0391889049599
Expected upper limit -2 sigma: sig_yield = 14.07008627303169
from utils import plotlimit
plotlimit(ul, CLs=False)
<Axes: xlabel='parameter of interest', ylabel='p-value'>
Splot#
This is now an demonstration of the sPlot algorithm, described in Pivk:2004ty.
If a data sample is populated by different sources of events, like signal and background, sPlot is able to unfold the contributions of the different sources for a given variable.
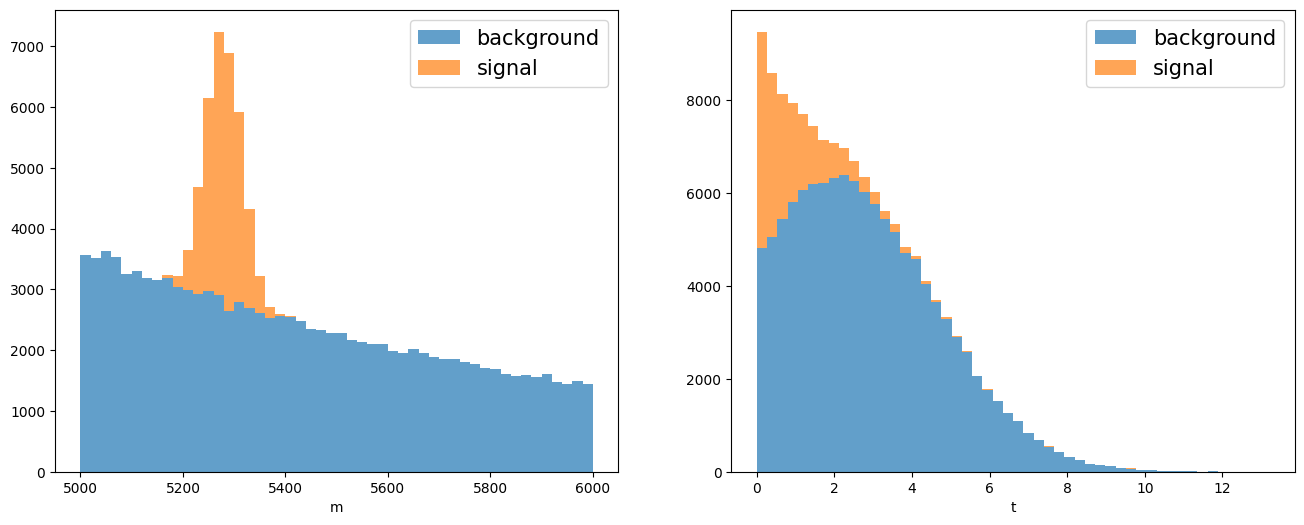
Let’s construct a dataset with two variables, the invariant mass and lifetime, for the resonant signal defined above and the combinatorial background.
# Signal distributions.
nsig_sw = 20000
np_sig_m_sw = signal_reso.sample(nsig_sw)["Bmass_reso"]
np_sig_t_sw = np.random.exponential(size=nsig_sw, scale=1)
# Background distributions.
nbkg_sw = 150000
np_bkg_m_sw = comb_bkg_reso.sample(nbkg_sw)["Bmass_reso"]
np_bkg_t_sw = np.random.normal(size=nbkg_sw, loc=2.0, scale=2.5)
# Lifetime cut.
t_cut = np_bkg_t_sw > 0
np_bkg_t_sw = np_bkg_t_sw[t_cut]
np_bkg_m_sw = np_bkg_m_sw[t_cut]
# Mass distribution
np_m_sw = np.concatenate([np_sig_m_sw, np_bkg_m_sw])
# Lifetime distribution
np_t_sw = np.concatenate([np_sig_t_sw, np_bkg_t_sw])
# Plots the mass and lifetime distribution.
fig, axs = plt.subplots(1, 2, figsize=(16, 6))
axs[0].hist([np_bkg_m_sw, np_sig_m_sw], bins=50, stacked=True, label=("background", "signal"), alpha=.7)
axs[0].set_xlabel("m")
axs[0].legend(fontsize=15)
axs[1].hist([np_bkg_t_sw, np_sig_t_sw], bins=50, stacked=True, label=("background", "signal"), alpha=.7)
axs[1].set_xlabel("t")
axs[1].legend(fontsize=15);
In this particular example we want to unfold the signal lifetime distribution. To do so sPlot needs a discriminant variable to determine the yields of the various sources using an extended maximum likelihood fit.
# Builds the loss.
data_sw = zfit.Data(np_m_sw, obs=obs_reso)
nll_sw = zfit.loss.ExtendedUnbinnedNLL(model_reso, data_sw)
#This parameter was useful in the simultaneous fit but not anymore so we fix it.
sigma_scaling.floating = False
# Minimizes the loss.
result_sw = minimizer.minimize(nll_sw)
print(result_sw.params)
name value (rounded) at limit
-------------- ------------------ ----------
reso_bkg_yield 117876 False
reso_sig_yield 20130 False
lambda_reso -0.000957066 False
mu 5278.99 False
sigma 40.8382 False
# Visualization of the result.
zfit.param.set_values(nll_sw.get_params(), result_sw)
plot_fit_projection(model_reso, data_sw, nbins=100)
<Axes: title={'center': 'Bmass_reso'}>
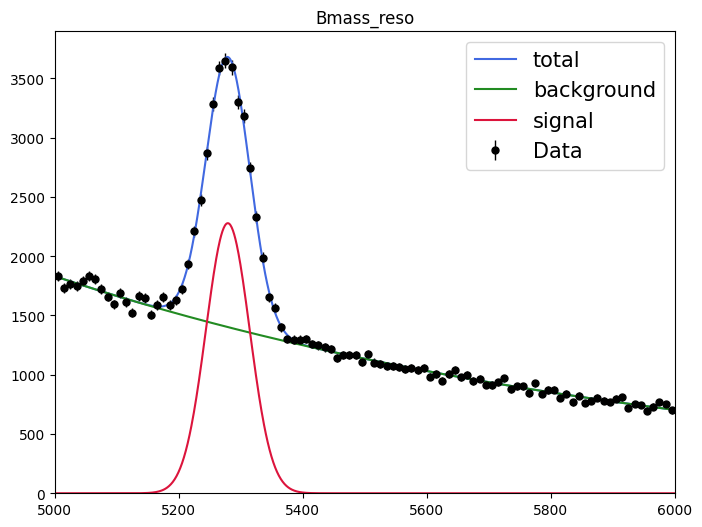
sPlot will use the fitted yield for each sources to derive the so-called sWeights for each data point:
with
where \({N_S}\) is the number of sources in the data sample, here 2. The index \(n\) represents the source, for instance \(0\) is the signal and \(1\) is the background, then \(f_0\) and \(N_0\) are the pdf and yield for the signal.
In hepstats the sWeights are computed with the compute_sweights function which takes as arguments the fitted extended model and the discrimant data (on which the fit was performed).
from hepstats.splot import compute_sweights
weights = compute_sweights(model_reso, data_sw)
print(weights)
{<zfit.Parameter 'reso_bkg_yield' floating=True value=1.179e+05>: array([-0.15627 , 0.30820479, -0.26438225, ..., 1.01417594,
1.19939028, 0.42460686]), <zfit.Parameter 'reso_sig_yield' floating=True value=2.013e+04>: array([ 1.15625313, 0.69178573, 1.26436367, ..., -0.01417418,
-0.19938558, 0.57538552])}
print("Sum of signal sWeights: ", np.sum(weights[reso_sig_yield]))
print("Sum of background sWeights: ", np.sum(weights[reso_bkg_yield]))
Sum of signal sWeights:
20130.005173097292
Sum of background sWeights:
117875.88586562786
Now we can apply the signal sWeights on the lifetime distribution and retrieve its signal components.
fig, axs = plt.subplots(1, 2, figsize=(16, 6))
nbins = 40
sorter = np_m_sw.argsort()
axs[0].plot(np_m_sw[sorter], weights[reso_sig_yield][sorter], label="$w_\\mathrm{sig}$")
axs[0].plot(np_m_sw[sorter], weights[reso_bkg_yield][sorter], label="$w_\\mathrm{bkg}$")
axs[0].plot(np_m_sw[sorter], weights[reso_sig_yield][sorter] + weights[reso_bkg_yield][sorter],
"-k", label="$w_\\mathrm{sig} + w_\\mathrm{bkg}$")
axs[0].axhline(0, color="0.5")
axs[0].legend(fontsize=15)
axs[0].set_xlim(5000, 5600)
axs[1].hist(np_t_sw, bins=nbins, range=(0, 6), weights=weights[reso_sig_yield], label="weighted histogram", alpha=.5)
axs[1].hist(np_sig_t_sw, bins=nbins, range=(0, 6), histtype="step", label="true histogram", lw=1.5)
axs[1].set_xlabel("t")
axs[1].legend(fontsize=15);
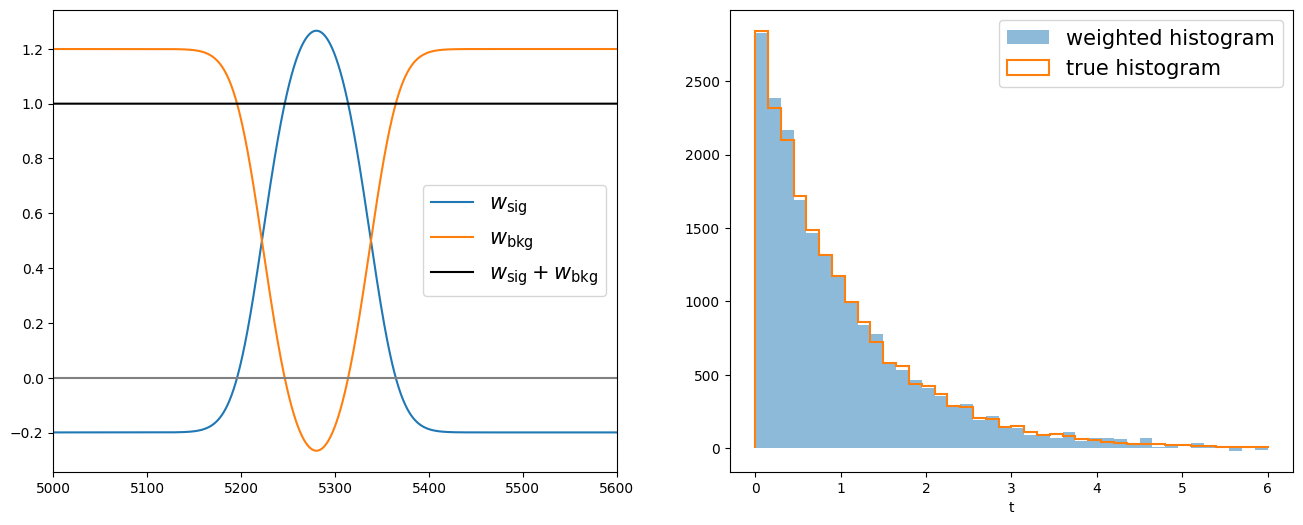
Be careful the sPlot technique works only on variables that are uncorrelated with the discriminant variable.
print(f"Correlation between m and t: {np.corrcoef(np_m_sw, np_t_sw)[0, 1]}")
Correlation between m and t: 0.06903206265730294
Let’s apply to signal sWeights on the mass distribution to see how bad the results of sPlot is when applied on a variable that is correlated with the discrimant variable.
plt.hist(np_m_sw, bins=100, range=(5000, 6000), weights=weights[reso_sig_yield]);