Show code cell content
%%capture
from IPython import get_ipython
install_packages = "google.colab" in str(get_ipython())
if install_packages:
!pip install zfit
Quickstart#
In this quick tutorial, we’ll show the basic ideas on what you can do with zfit, without going into much detail or performing advanced tasks.
import numpy as np
import zfit
import zfit.z.numpy # numpy-like backend
from zfit import z # backend
No module named 'nlopt'
Create observables#
The observable space in which PDFs are defined is created with the Space class
obs = zfit.Space('x', limits=(-10, 10))
Create data#
We create some unbinned data using numpy. Other constructors, e.g. for ROOT files are also available.
mu_true = 0
sigma_true = 1
data_np = np.random.normal(mu_true, sigma_true, size=10000)
data = zfit.data.Data.from_numpy(obs=obs, array=data_np)
Create a PDF to fit#
Let’s create a Gaussian PDF so we can fit the dataset. To do this, first we create the fit parameters, which follow a convention similar to RooFit:
zfit.Parameter(name, initial_value, lower_limit (optional), upper_limit (optional), other options)
mu = zfit.Parameter("mu", 2.4, -1., 5., step_size=0.001) # step_size is not mandatory but can be helpful
sigma = zfit.Parameter("sigma", 1.3, 0, 5., step_size=0.001) # it should be around the estimated uncertainty
Now we instantiate a Gaussian from the zfit PDF library (more on how to create your own PDFs later)
gauss = zfit.pdf.Gauss(obs=obs, mu=mu, sigma=sigma)
This pdf contains several useful methods, such as calculating a probability, calculating its integral, sampling etc.
Note: Several objects that are returned from methods, like integrate, return tf.Tensor, which are wrapped Numpy arrays.
They can directly be used as such or explicitly converted to by calling:
zfit.run(TensorFlow_object)
# Let's get some probabilities.
consts = [-1, 0, 1]
probs = gauss.pdf(consts)
# And now execute the tensorflow graph
result = zfit.run(probs)
print(f"x values: {consts}\nresult: {result}")
x values: [-1, 0, 1]
result: [0.01003756 0.05582994 0.17184119]
Fitting#
To fit, we need to take three steps: create the negative \(\log\mathcal{L}\), instantiate a minimizer and then minimize the likelihood.
# Create the negative log likelihood
nll = zfit.loss.UnbinnedNLL(model=gauss, data=data) # loss
# Load and instantiate a minimizer
minimizer = zfit.minimize.Minuit()
minimum = minimizer.minimize(loss=nll)
params = minimum.params
print(minimum)
FitResult
of
<UnbinnedNLL model=[<zfit.<class 'zfit.models.dist_tfp.Gauss'> params=[mu, sigma]] data=[<zfit.core.data.Data object at 0x7f544c3a1150>] constraints=[]>
with
<Minuit Minuit tol=0.001>
╒═════════╤═════════════╤══════════════════╤═════════╤══════════════════════════════════╕
│ valid │ converged │ param at limit │ edm │ approx. fmin (full | internal) │
╞═════════╪═════════════╪══════════════════╪═════════╪══════════════════════════════════╡
│
True
│ True
│ False
│ 1.1e-05 │ 14193.08 | -7712.851 │
╘═════════╧═════════════╧══════════════════╧═════════╧══════════════════════════════════╛
Parameters
name value (rounded) at limit
------ ------------------ ----------
mu -0.00640465 False
sigma 1.0004 False
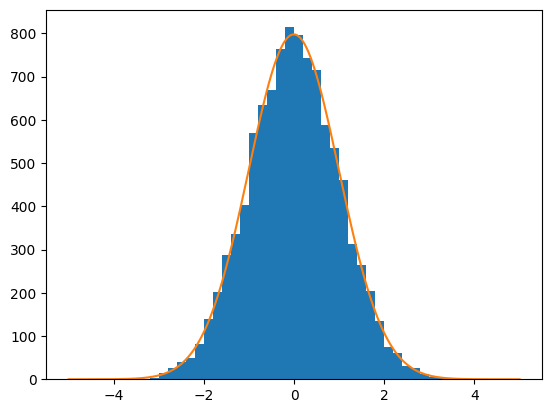
And we can plot the result to see how it went.
%matplotlib inline
import matplotlib.pyplot as plt
n_bins = 50
range_ = (-5,5)
_ = plt.hist(data_np, bins=n_bins, range=range_)
x = np.linspace(*range_, num=1000)
pdf = zfit.run(gauss.pdf(x))
_ = plt.plot(x, data_np.shape[0] / n_bins * (range_[1] - range_[0]) * pdf)
The FitResult that we obtained contains information about the minimization and can now be used to calculate the errors
print(f"Function minimum: {minimum.fmin}", minimum.fmin)
print(f"Converged: {minimum.converged} and valid: {minimum.valid}", )
print(minimum)
Function minimum: -7712.850874254467
-7712.850874254467
Converged: True and valid: True
FitResult
of
<UnbinnedNLL model=[<zfit.<class 'zfit.models.dist_tfp.Gauss'> params=[mu, sigma]] data=[<zfit.core.data.Data object at 0x7f544c3a1150>] constraints=[]>
with
<Minuit Minuit tol=0.001>
╒═════════╤═════════════╤══════════════════╤═════════╤══════════════════════════════════╕
│ valid │ converged │ param at limit │ edm │ approx. fmin (full | internal) │
╞═════════╪═════════════╪══════════════════╪═════════╪══════════════════════════════════╡
│
True
│ True
│ False
│ 1.1e-05 │ 14193.08 | -7712.851 │
╘═════════╧═════════════╧══════════════════╧═════════╧══════════════════════════════════╛
Parameters
name value (rounded) at limit
------ ------------------ ----------
mu -0.00640465 False
sigma 1.0004 False
hesse_errors = minimum.hesse()
print(minimum)
FitResult
of
<UnbinnedNLL model=[<zfit.<class 'zfit.models.dist_tfp.Gauss'> params=[mu, sigma]] data=[<zfit.core.data.Data object at 0x7f544c3a1150>] constraints=[]>
with
<Minuit Minuit tol=0.001>
╒═════════╤═════════════╤══════════════════╤═════════╤══════════════════════════════════╕
│ valid │ converged │ param at limit │ edm │ approx. fmin (full | internal) │
╞═════════╪═════════════╪══════════════════╪═════════╪══════════════════════════════════╡
│
True
│ True
│ False
│ 1.1e-05 │ 14193.08 | -7712.851 │
╘═════════╧═════════════╧══════════════════╧═════════╧══════════════════════════════════╛
Parameters
name value (rounded) hesse at limit
------ ------------------ ----------- ----------
mu -0.00640465 +/- 0.01 False
sigma 1.0004 +/- 0.0071 False


